Directed evolution is the process of mutagenizing and screening biomolecules to select specific desired properties, or even unprecedented properties. Directed evolution allows researchers to address increasingly complex questions by creating biomolecules useful in treating disease, removing plastic, carbon, or carbon dioxide from the environment, and synthesizing novel complex organic molecules. Conventional directed evolution methods are laborious and time-consuming because only relatively short genetic segments can be evolved at one time and are limited to a single biomolecule. Extra molecular biology cloning steps may cause errors or loss of the DNA library.
GW researchers developed a novel method for the directed evolution of biomolecules using a single PCR mammalian expression generated array (MEGA). This method is rapid and requires less hands-on experimental time than conventional methods. The new procedure uses error prone PCR amplification of an entire plasmid (> 7 kilobases), enabling directed evolution of multiple protein genes, DNA, or RNA to be evolved together for synthetic biology applications. E. coli circularizes the linear DNA for biomolecule(s) expression in mammalian cells. Circular DNA is purified using standard kits after selection.
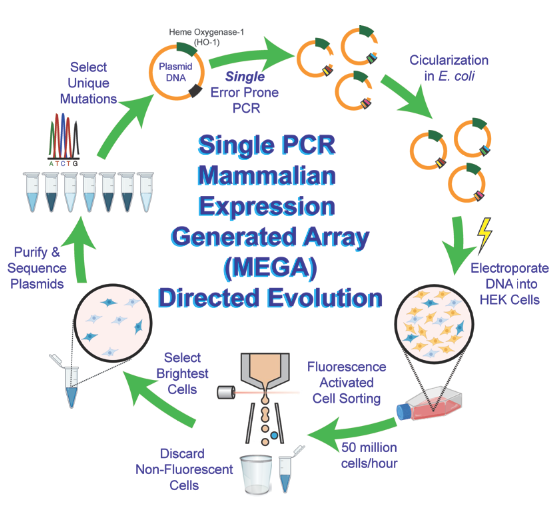
Figure. Single PCR MEGA Directed Evolution of Biomolecules schematic. Shown is a fluorescent protein (blue) evolution with high-throughput screening by fluorescence.
Applications:
- Evolution of novel biomolecules to select for new or improved functions
- Applicable to proteins, antibodies, enzymes, RNA, and DNA
Advantages:
- Removes most molecular biology, faster (3 hours), and safer than viral methods
- High mutation efficiency
- Evolve multiple biomolecules on a single plasmid
- Works with any cell type that can be genetically modified